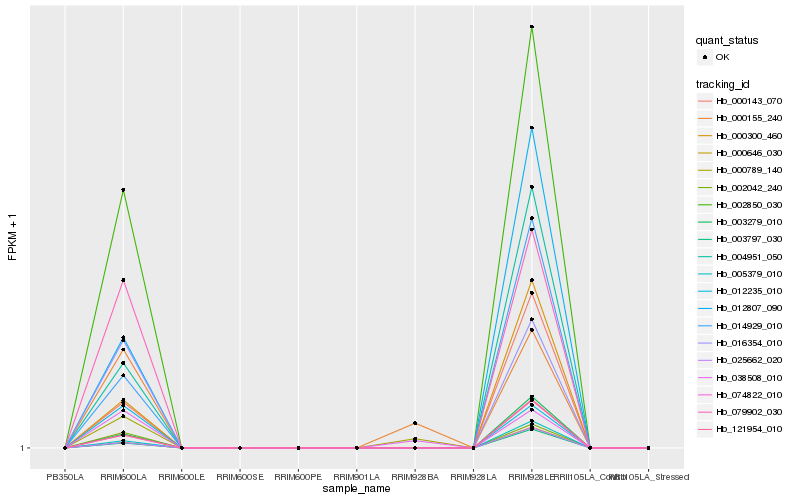
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_079902_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_016354_010 |
0.0213210744 |
- |
- |
- |
| 3 |
Hb_002042_240 |
0.0308326874 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_012235_010 |
0.0571980099 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 5 |
Hb_002850_030 |
0.0575820501 |
- |
- |
- |
| 6 |
Hb_012807_090 |
0.1717999897 |
- |
- |
PREDICTED: uncharacterized protein LOC100267173 [Vitis vinifera] |
| 7 |
Hb_004951_050 |
0.1794324885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_014929_010 |
0.1831491011 |
- |
- |
- |
| 9 |
Hb_000143_070 |
0.1912594606 |
- |
- |
- |
| 10 |
Hb_000300_460 |
0.1978361333 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 11 |
Hb_003279_010 |
0.2009942875 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 12 |
Hb_121954_010 |
0.2012590864 |
- |
- |
PREDICTED: uncharacterized protein LOC105774125 [Gossypium raimondii] |
| 13 |
Hb_074822_010 |
0.2013347799 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 14 |
Hb_005379_010 |
0.2030676522 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_000646_030 |
0.2035879992 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_025662_020 |
0.2036204551 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_003797_030 |
0.2037542401 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_038508_010 |
0.2183833545 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 19 |
Hb_000155_240 |
0.228045416 |
- |
- |
PREDICTED: uncharacterized protein LOC104888642 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000789_140 |
0.2321192278 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |