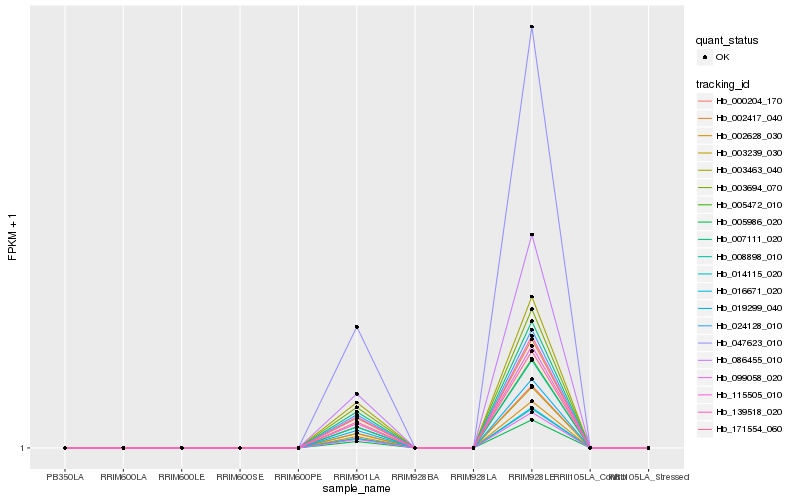
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016671_020 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_014115_020 |
0.0003234727 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 3 |
Hb_099058_020 |
0.0012608723 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_002628_030 |
0.0017114902 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_003239_030 |
0.002579099 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_086455_010 |
0.0028088057 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 7 |
Hb_008898_010 |
0.0032825579 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_115505_010 |
0.0042891957 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_005472_010 |
0.0050633916 |
- |
- |
- |
| 10 |
Hb_000204_170 |
0.0066332232 |
- |
- |
integrase [Gossypium herbaceum] |
| 11 |
Hb_019299_040 |
0.0078775043 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor ABI3 [Jatropha curcas] |
| 12 |
Hb_005986_020 |
0.0085940597 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 13 |
Hb_171554_060 |
0.0162213541 |
- |
- |
PREDICTED: uncharacterized protein LOC102627687 [Citrus sinensis] |
| 14 |
Hb_047623_010 |
0.0183876763 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 15 |
Hb_002417_040 |
0.0199721691 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 16 |
Hb_139518_020 |
0.0210331432 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_024128_010 |
0.0231040934 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |
| 18 |
Hb_007111_020 |
0.0259160251 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 19 |
Hb_003694_070 |
0.0300724793 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 20 |
Hb_003463_040 |
0.034547003 |
- |
- |
- |