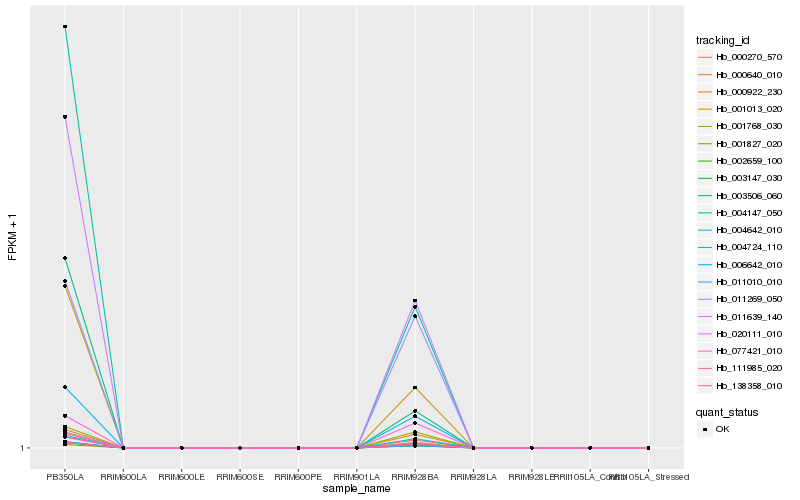
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004147_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_03640 [Jatropha curcas] |
| 2 |
Hb_020111_010 |
0.0041423818 |
- |
- |
PREDICTED: uncharacterized protein LOC102661177 [Glycine max] |
| 3 |
Hb_001013_020 |
0.0540300454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003506_060 |
0.0629618454 |
- |
- |
PREDICTED: glutathione S-transferase F9-like [Jatropha curcas] |
| 5 |
Hb_011010_010 |
0.0694757147 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_011639_140 |
0.0698195025 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 7 |
Hb_000640_010 |
0.0717027694 |
- |
- |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 8 |
Hb_002659_100 |
0.0758518647 |
- |
- |
hypothetical protein OsI_37193 [Oryza sativa Indica Group] |
| 9 |
Hb_111985_020 |
0.0773548546 |
- |
- |
- |
| 10 |
Hb_006642_010 |
0.1327887216 |
- |
- |
- |
| 11 |
Hb_003147_030 |
0.2013940103 |
transcription factor |
TF Family: Jumonji |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_138358_010 |
0.2016231979 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 13 |
Hb_004642_010 |
0.2019789534 |
- |
- |
Integrase core domain, putative [Oryza sativa Japonica Group] |
| 14 |
Hb_000270_570 |
0.2044786364 |
- |
- |
PREDICTED: peroxidase 65-like [Jatropha curcas] |
| 15 |
Hb_004724_110 |
0.2054560022 |
- |
- |
alpha-1,2-fucosyltransferase [Populus tremula x Populus alba] |
| 16 |
Hb_000922_230 |
0.2087551039 |
- |
- |
- |
| 17 |
Hb_001768_030 |
0.2104932724 |
- |
- |
- |
| 18 |
Hb_001827_020 |
0.2143642729 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_011269_050 |
0.2151290867 |
- |
- |
- |
| 20 |
Hb_077421_010 |
0.2169829242 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |