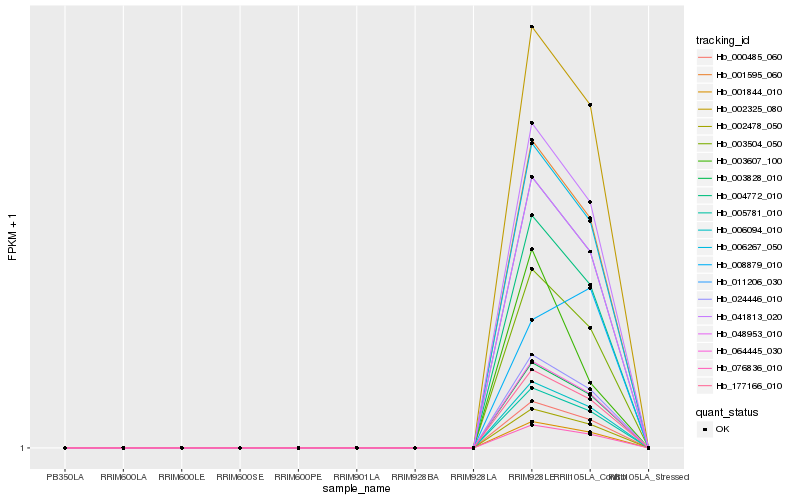
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003504_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_23540 [Jatropha curcas] |
| 2 |
Hb_004772_010 |
0.0082470892 |
- |
- |
PREDICTED: uncharacterized protein LOC104647507 [Solanum lycopersicum] |
| 3 |
Hb_024446_010 |
0.0120395953 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 4 |
Hb_064445_030 |
0.0128913038 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 5 |
Hb_003828_010 |
0.0130882937 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 6 |
Hb_177166_010 |
0.0139968428 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 7 |
Hb_011206_030 |
0.014390914 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_048953_010 |
0.014390914 |
- |
- |
PREDICTED: uncharacterized protein LOC105775428 [Gossypium raimondii] |
| 9 |
Hb_006094_010 |
0.0155261487 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 10 |
Hb_005781_010 |
0.0163095696 |
- |
- |
PREDICTED: probable terpene synthase 6 [Jatropha curcas] |
| 11 |
Hb_000485_060 |
0.0179859876 |
- |
- |
PREDICTED: uncharacterized protein LOC105641351 [Jatropha curcas] |
| 12 |
Hb_002478_050 |
0.0188968448 |
- |
- |
K(+)/H(+) antiporter, putative [Ricinus communis] |
| 13 |
Hb_006267_050 |
0.0198401312 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 14 |
Hb_001595_060 |
0.020357969 |
- |
- |
hypothetical protein L484_001429 [Morus notabilis] |
| 15 |
Hb_001844_010 |
0.0204408762 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 16 |
Hb_076836_010 |
0.0208396677 |
- |
- |
PREDICTED: uncharacterized protein LOC104801925 [Tarenaya hassleriana] |
| 17 |
Hb_041813_020 |
0.0231186692 |
- |
- |
ATP synthase epsilon subunit [Grubbia rosmarinifolia] |
| 18 |
Hb_002325_080 |
0.03855133 |
- |
- |
- |
| 19 |
Hb_008879_010 |
0.1385656563 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 20 |
Hb_003607_100 |
0.1453938022 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |