| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
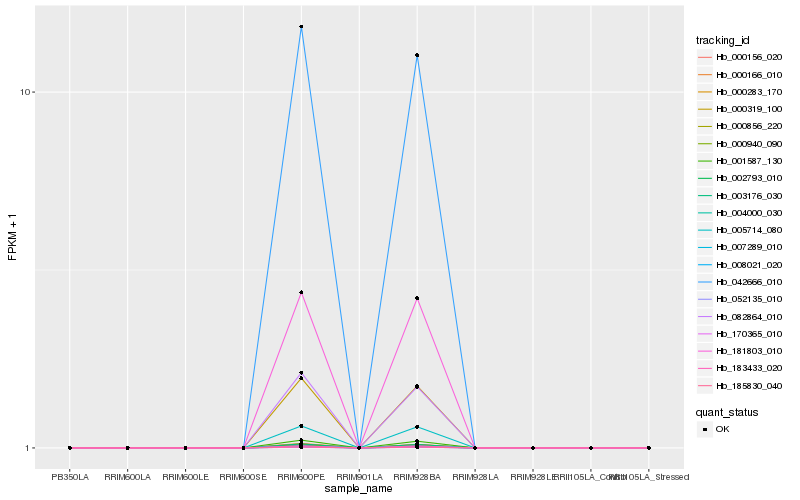
Hb_001587_130 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 2 |
Hb_000319_100 |
0.0003341438 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 3 |
Hb_000156_020 |
0.0010680523 |
- |
- |
PREDICTED: la-related protein 6A [Jatropha curcas] |
| 4 |
Hb_003176_030 |
0.0010680523 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Solanum lycopersicum] |
| 5 |
Hb_000940_090 |
0.0029177499 |
- |
- |
hypothetical protein CISIN_1g046520mg [Citrus sinensis] |
| 6 |
Hb_000283_170 |
0.0087222442 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_008021_020 |
0.0090359256 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0014s00780g [Populus trichocarpa] |
| 8 |
Hb_007289_010 |
0.0091881013 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_042666_010 |
0.0108233301 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000856_220 |
0.0164894735 |
- |
- |
hypothetical protein POPTR_0012s07300g [Populus trichocarpa] |
| 11 |
Hb_002793_010 |
0.0169094497 |
- |
- |
putative metal-nicotianamine transporter YSL7 -like protein [Gossypium arboreum] |
| 12 |
Hb_185830_040 |
0.0173561806 |
- |
- |
hypothetical protein JCGZ_16423 [Jatropha curcas] |
| 13 |
Hb_004000_030 |
0.018522106 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 14 |
Hb_181803_010 |
0.0185959035 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 15 |
Hb_183433_020 |
0.0198034322 |
- |
- |
PREDICTED: uncharacterized protein LOC105851424 [Cicer arietinum] |
| 16 |
Hb_170365_010 |
0.0204046672 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 17 |
Hb_052135_010 |
0.0205045847 |
- |
- |
PREDICTED: phytochrome B isoform X2 [Jatropha curcas] |
| 18 |
Hb_005714_080 |
0.0222806163 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 19 |
Hb_082864_010 |
0.0227057001 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 20 |
Hb_000166_010 |
0.0229584415 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |