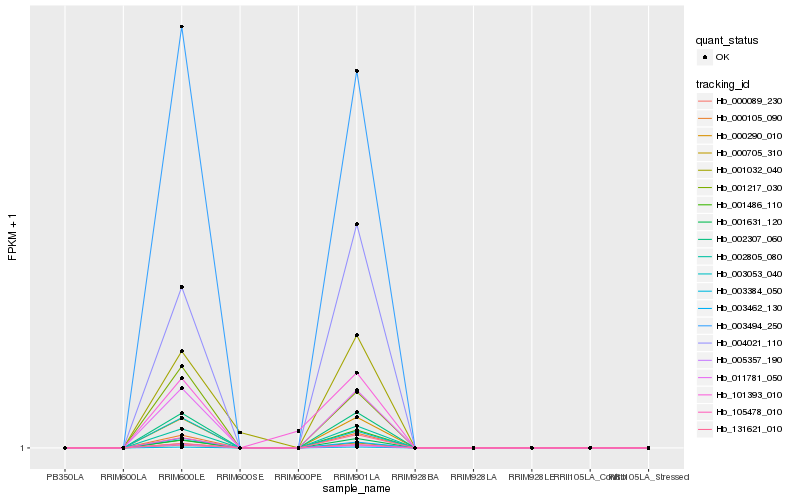
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000290_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002307_060 |
0.0029717921 |
- |
- |
- |
| 3 |
Hb_000089_230 |
0.0149992188 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 4 |
Hb_003384_050 |
0.016084682 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_011781_050 |
0.0162891589 |
- |
- |
- |
| 6 |
Hb_002805_080 |
0.0219857863 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 7 |
Hb_000105_090 |
0.0237448034 |
- |
- |
- |
| 8 |
Hb_001631_120 |
0.0249961282 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000705_310 |
0.026008401 |
- |
- |
hypothetical protein POPTR_0001s03390g [Populus trichocarpa] |
| 10 |
Hb_003494_250 |
0.056255984 |
- |
- |
- |
| 11 |
Hb_004021_110 |
0.0915807772 |
- |
- |
- |
| 12 |
Hb_001217_030 |
0.0984411427 |
- |
- |
- |
| 13 |
Hb_005357_190 |
0.099301266 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 14 |
Hb_003462_130 |
0.1197960055 |
- |
- |
- |
| 15 |
Hb_003053_040 |
0.1368807965 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001486_110 |
0.1674131025 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 17 |
Hb_001032_040 |
0.1740727195 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_131621_010 |
0.215326825 |
- |
- |
hypothetical protein MIMGU_mgv1a004274mg [Erythranthe guttata] |
| 19 |
Hb_101393_010 |
0.221285373 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_105478_010 |
0.2338582036 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |