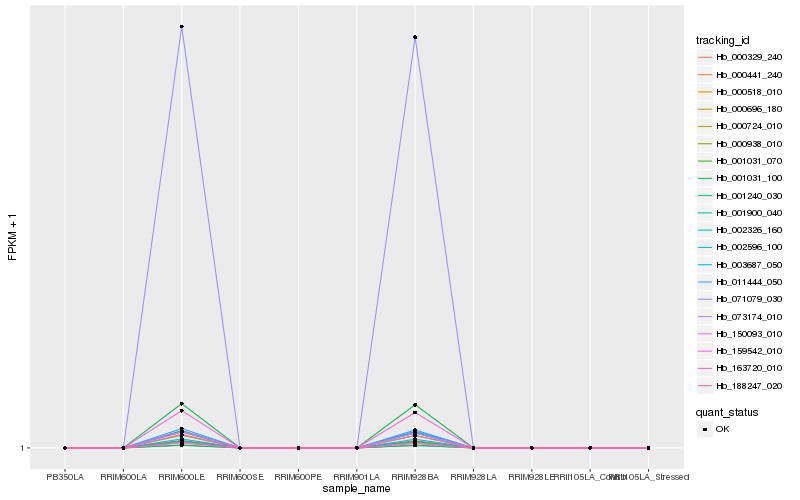
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163720_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 2 |
Hb_073174_010 |
0.0007310803 |
- |
- |
PREDICTED: MLP-like protein 328 [Jatropha curcas] |
| 3 |
Hb_000329_240 |
0.0009685383 |
transcription factor |
TF Family: PLATZ |
zinc ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_011444_050 |
0.0014834898 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 5 |
Hb_001900_040 |
0.0015789172 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 6 |
Hb_000518_010 |
0.0017014543 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17300g [Populus trichocarpa] |
| 7 |
Hb_188247_020 |
0.0019166109 |
- |
- |
hypothetical protein, partial [Cecembia lonarensis] |
| 8 |
Hb_150093_010 |
0.0021430152 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 9 |
Hb_002326_160 |
0.0023174981 |
- |
- |
hypothetical protein JCGZ_04332 [Jatropha curcas] |
| 10 |
Hb_001240_030 |
0.0026866597 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 11 |
Hb_000696_180 |
0.004485473 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 12 |
Hb_159542_010 |
0.0059352178 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 13 |
Hb_000938_010 |
0.0084548805 |
- |
- |
polyprotein [Oryza australiensis] |
| 14 |
Hb_071079_030 |
0.0097065927 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 15 |
Hb_001031_070 |
0.0100945232 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 16 |
Hb_000441_240 |
0.0102863669 |
- |
- |
hypothetical protein ZEAMMB73_005804 [Zea mays] |
| 17 |
Hb_003687_050 |
0.0108304566 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002596_100 |
0.0109686249 |
- |
- |
- |
| 19 |
Hb_001031_100 |
0.0120217298 |
- |
- |
- |
| 20 |
Hb_000724_010 |
0.0125374432 |
- |
- |
PREDICTED: uncharacterized protein LOC105134348 [Populus euphratica] |