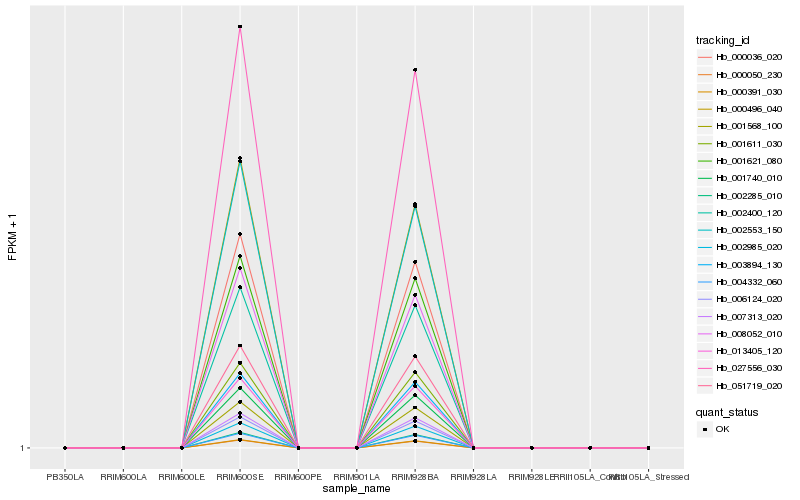
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007313_020 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_004332_060 |
0.0002342378 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000036_020 |
0.0006121811 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_002985_020 |
0.0006418854 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 5 |
Hb_000391_030 |
0.0007579052 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_000050_230 |
0.0007912051 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_002285_010 |
0.0008013583 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_006124_020 |
0.0011278464 |
- |
- |
hypothetical protein JCGZ_13738 [Jatropha curcas] |
| 9 |
Hb_001568_100 |
0.0024118251 |
- |
- |
hypothetical protein Csa_2G404850 [Cucumis sativus] |
| 10 |
Hb_001740_010 |
0.0036019945 |
- |
- |
hypothetical protein JCGZ_18625 [Jatropha curcas] |
| 11 |
Hb_008052_010 |
0.004083733 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001621_080 |
0.0041056089 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 13 |
Hb_013405_120 |
0.0044590311 |
- |
- |
- |
| 14 |
Hb_003894_130 |
0.0048605447 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002400_120 |
0.005486994 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 16 |
Hb_001611_030 |
0.0057862333 |
- |
- |
- |
| 17 |
Hb_027556_030 |
0.0063219203 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like, partial [Gossypium raimondii] |
| 18 |
Hb_002553_150 |
0.0071267084 |
- |
- |
Gram-positive signal peptide protein, YSIRK family [Streptococcus pneumoniae TCH8431/19A] |
| 19 |
Hb_051719_020 |
0.0072884954 |
- |
- |
PREDICTED: toll/interleukin-1 receptor-like protein [Jatropha curcas] |
| 20 |
Hb_000496_040 |
0.007587448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |