| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
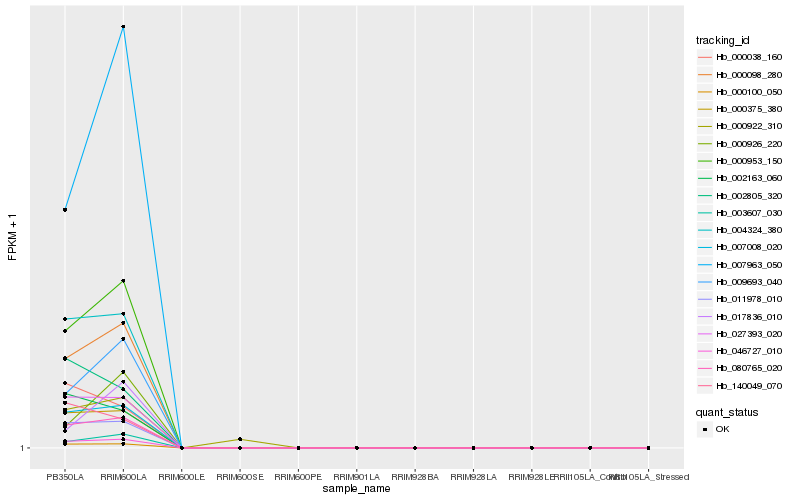
Hb_000953_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_000098_280 |
0.0066537104 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g33350 [Jatropha curcas] |
| 3 |
Hb_080765_020 |
0.0208705128 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_046727_010 |
0.0240579626 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_007008_020 |
0.0444827903 |
- |
- |
- |
| 6 |
Hb_007963_050 |
0.0654904479 |
- |
- |
- |
| 7 |
Hb_000100_050 |
0.0695174105 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 8 |
Hb_011978_010 |
0.0702547425 |
- |
- |
hypothetical protein JCGZ_03852 [Jatropha curcas] |
| 9 |
Hb_000375_380 |
0.0706132453 |
- |
- |
- |
| 10 |
Hb_009693_040 |
0.072178991 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070 [Brassica rapa] |
| 11 |
Hb_004324_380 |
0.0740793569 |
- |
- |
- |
| 12 |
Hb_003607_030 |
0.0827689157 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor ABI3 [Jatropha curcas] |
| 13 |
Hb_027393_020 |
0.0860006919 |
- |
- |
- |
| 14 |
Hb_002163_060 |
0.1648652734 |
- |
- |
- |
| 15 |
Hb_002805_320 |
0.1770833879 |
- |
- |
- |
| 16 |
Hb_000926_220 |
0.1780401957 |
- |
- |
- |
| 17 |
Hb_140049_070 |
0.1810454988 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000038_160 |
0.1820802296 |
- |
- |
putative gag protein [Coffea canephora] |
| 19 |
Hb_017836_010 |
0.189134632 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 20 |
Hb_000922_310 |
0.2125899567 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB98-like [Jatropha curcas] |