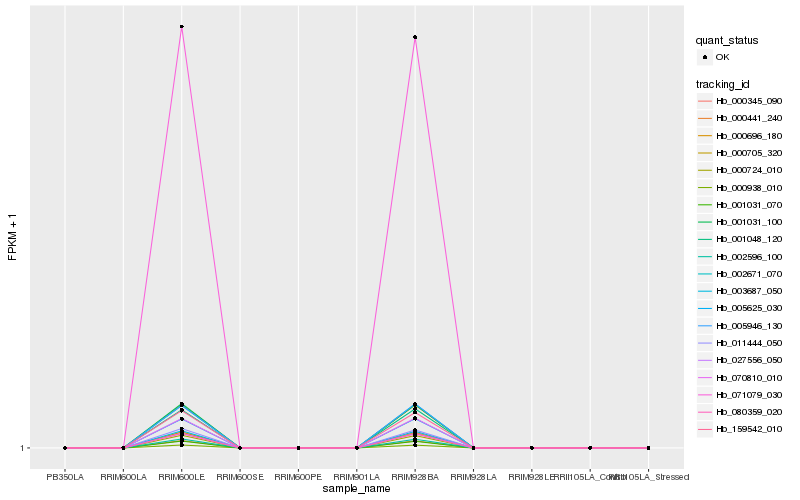
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_320 |
0.0 |
- |
- |
Transcriptional regulator SUPERMAN, putative [Ricinus communis] |
| 2 |
Hb_000345_090 |
0.0026895241 |
- |
- |
Cell cycle checkpoint control protein family isoform 5 [Theobroma cacao] |
| 3 |
Hb_005625_030 |
0.0030549515 |
- |
- |
ribosomal protein S12 (mitochondrion) [Chrysobalanus icaco] |
| 4 |
Hb_027556_050 |
0.0031906762 |
- |
- |
hypothetical protein PHAVU_001G166100g [Phaseolus vulgaris] |
| 5 |
Hb_070810_010 |
0.0032240517 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 6 |
Hb_080359_020 |
0.0033552523 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 7 |
Hb_000724_010 |
0.0040593927 |
- |
- |
PREDICTED: uncharacterized protein LOC105134348 [Populus euphratica] |
| 8 |
Hb_001031_100 |
0.0045751297 |
- |
- |
- |
| 9 |
Hb_002596_100 |
0.0056282735 |
- |
- |
- |
| 10 |
Hb_003687_050 |
0.0057664459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000441_240 |
0.0063105498 |
- |
- |
hypothetical protein ZEAMMB73_005804 [Zea mays] |
| 12 |
Hb_001031_070 |
0.0065023977 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 13 |
Hb_071079_030 |
0.0068903353 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 14 |
Hb_001048_120 |
0.0069303611 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 15 |
Hb_000938_010 |
0.0081420589 |
- |
- |
polyprotein [Oryza australiensis] |
| 16 |
Hb_002671_070 |
0.0087674867 |
- |
- |
hypothetical protein POPTR_0006s05220g, partial [Populus trichocarpa] |
| 17 |
Hb_005946_130 |
0.0087674867 |
- |
- |
- |
| 18 |
Hb_159542_010 |
0.0106616896 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_000696_180 |
0.0121113829 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_011444_050 |
0.0151131823 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |