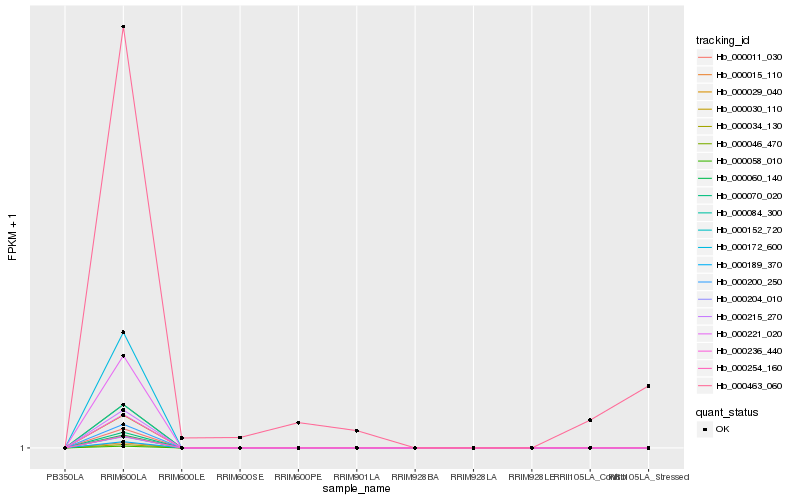
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000463_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 2 |
Hb_000011_030 |
0.2857860476 |
- |
- |
hypothetical protein POPTR_0003s18920g [Populus trichocarpa] |
| 3 |
Hb_000015_110 |
0.2857860476 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 4 |
Hb_000029_040 |
0.2857860476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000030_110 |
0.2857860476 |
- |
- |
- |
| 6 |
Hb_000034_130 |
0.2857860476 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000046_470 |
0.2857860476 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 8 |
Hb_000058_010 |
0.2857860476 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000060_140 |
0.2857860476 |
- |
- |
- |
| 10 |
Hb_000070_020 |
0.2857860476 |
- |
- |
- |
| 11 |
Hb_000084_300 |
0.2857860476 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 12 |
Hb_000152_720 |
0.2857860476 |
- |
- |
- |
| 13 |
Hb_000172_600 |
0.2857860476 |
- |
- |
- |
| 14 |
Hb_000189_370 |
0.2857860476 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 15 |
Hb_000200_250 |
0.2857860476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000204_010 |
0.2857860476 |
- |
- |
hypothetical protein POPTR_0004s13450g [Populus trichocarpa] |
| 17 |
Hb_000215_270 |
0.2857860476 |
- |
- |
- |
| 18 |
Hb_000221_020 |
0.2857860476 |
- |
- |
- |
| 19 |
Hb_000236_440 |
0.2857860476 |
- |
- |
- |
| 20 |
Hb_000254_160 |
0.2857860476 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |