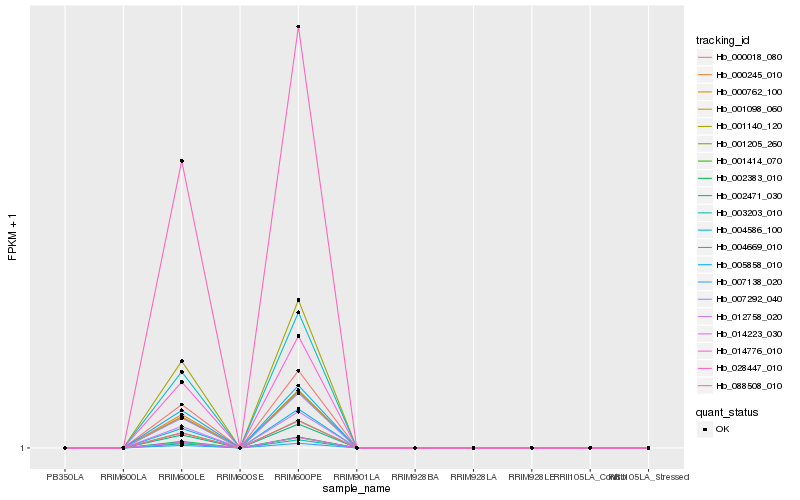
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_07748 [Jatropha curcas] |
| 2 |
Hb_007292_040 |
0.0021178953 |
- |
- |
Plastid developmental protein DAG [Theobroma cacao] |
| 3 |
Hb_000762_100 |
0.0025476069 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |
| 4 |
Hb_001140_120 |
0.0028504759 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 5 |
Hb_002471_030 |
0.0030092051 |
- |
- |
PREDICTED: cyclin-J18 [Jatropha curcas] |
| 6 |
Hb_014223_030 |
0.003436303 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At4g03220 [Jatropha curcas] |
| 7 |
Hb_001098_060 |
0.0037106043 |
- |
- |
- |
| 8 |
Hb_088508_010 |
0.0042056103 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_014776_010 |
0.0047423811 |
- |
- |
PREDICTED: receptor-like protein kinase 5 [Jatropha curcas] |
| 10 |
Hb_007138_020 |
0.0056916784 |
- |
- |
hypothetical protein POPTR_0019s09630g [Populus trichocarpa] |
| 11 |
Hb_001414_070 |
0.0058690535 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 12 |
Hb_003203_010 |
0.0063177665 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 13 |
Hb_004586_100 |
0.0108493035 |
- |
- |
PREDICTED: uncharacterized protein LOC105120428 isoform X2 [Populus euphratica] |
| 14 |
Hb_000245_010 |
0.0112461082 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Populus euphratica] |
| 15 |
Hb_028447_010 |
0.0113361726 |
- |
- |
hypothetical protein RCOM_1967580 [Ricinus communis] |
| 16 |
Hb_001205_260 |
0.012900187 |
- |
- |
DWNN domain isoform 2 [Theobroma cacao] |
| 17 |
Hb_004669_010 |
0.0162547944 |
- |
- |
hypothetical protein JCGZ_26481 [Jatropha curcas] |
| 18 |
Hb_005858_010 |
0.0178054728 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_12260 [Jatropha curcas] |
| 19 |
Hb_012758_020 |
0.0197309608 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 20 |
Hb_002383_010 |
0.0240545688 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |